很久之前整理的关于R绘制维恩图的资料,虽然,用R绘制维恩图并不是很常见,还是记下一笔,以备需要者参考。
# Gplots包
library(gplots)
##
## 载入程序包:'gplots'
## The following object is masked from 'package:stats':
##
## lowess
A<- 1:20
B<- 1:20
C<- 2:20
D<- 3:21
input<-list(A,B,C,D)
input
## [[1]]
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
##
## [[2]]
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
##
## [[3]]
## [1] 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
##
## [[4]]
## [1] 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21
venn(input)
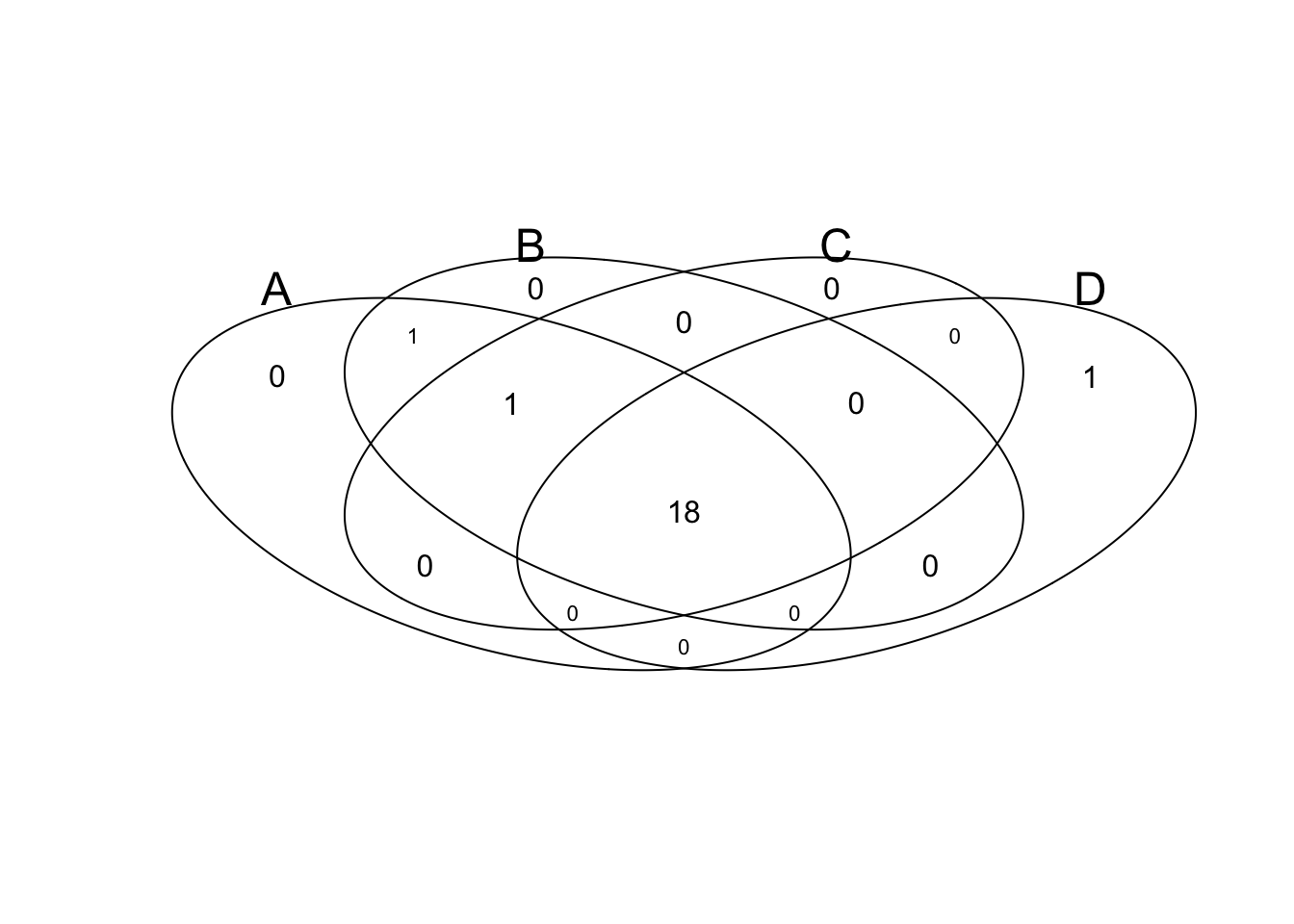
A<- as.logical(rbinom(100, 1, 0.2))
B<- as.logical(rbinom(100, 1, 0.7))
C<- as.logical(rbinom(100, 1, 0.2))
D<- as.logical(rbinom(100, 1, 0.1))
input<-data.frame(A,B,C,D)
venn(input)

tmp <- venn(input, simplify=TRUE)
## Warning in drawVennDiagram(data = counts, small = small, showSetLogicLabel = showSetLogicLabel, : Not shown: 0101 contains 2
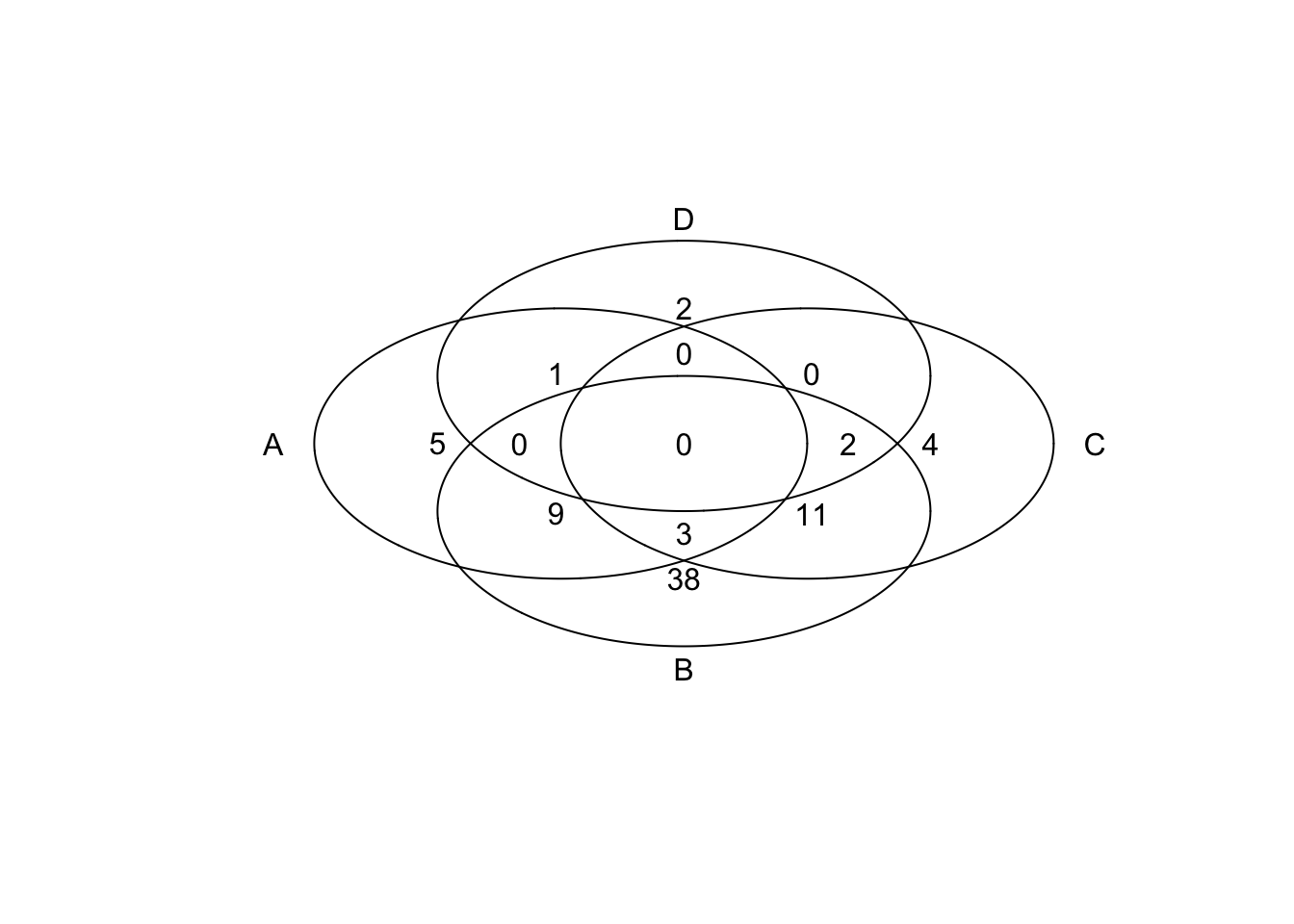
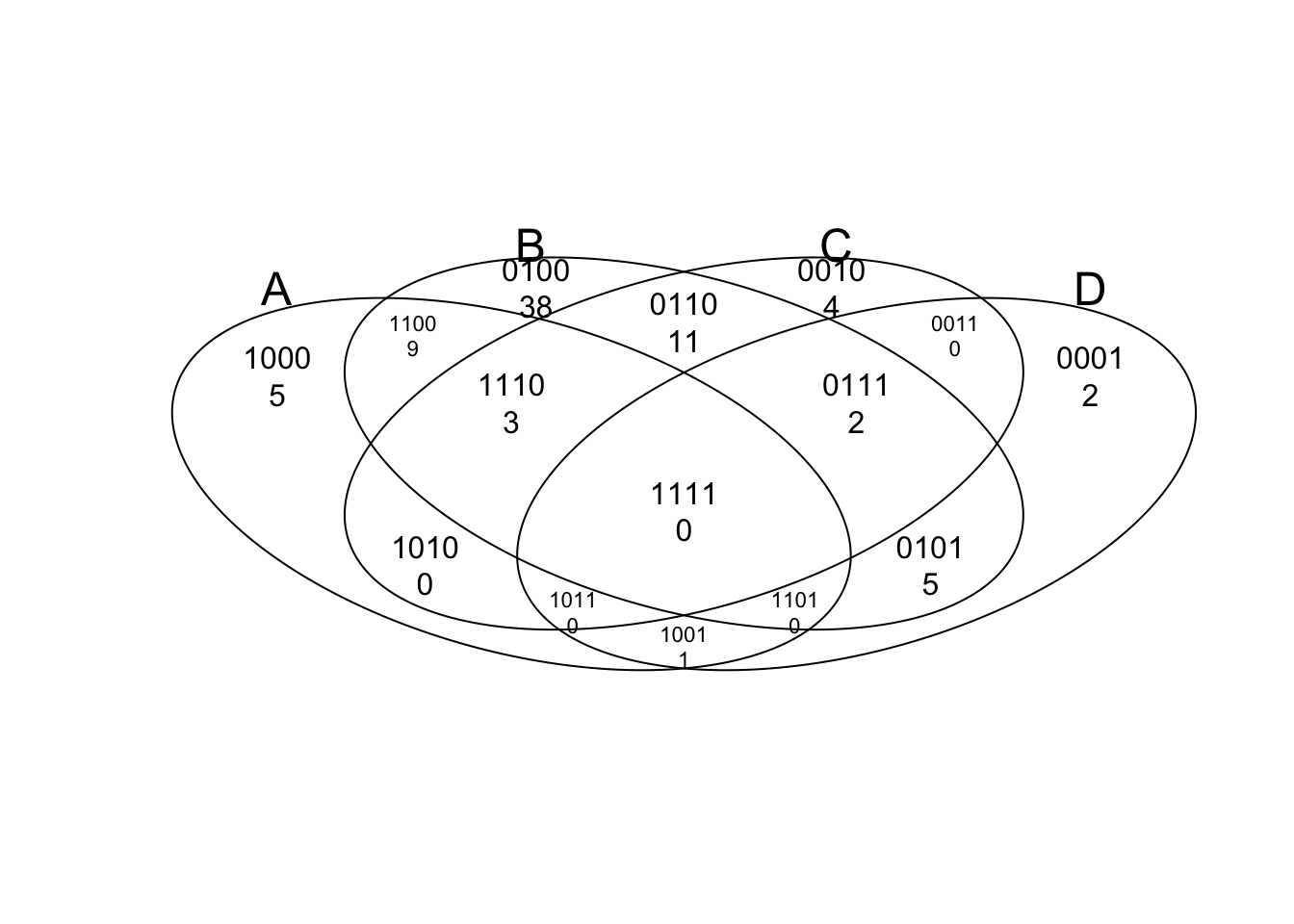
tmp
## num A B C D
## 0000 0 0 0 0 0
## 0001 1 0 0 0 1
## 0010 4 0 0 1 0
## 0011 0 0 0 1 1
## 0100 36 0 1 0 0
## 0101 2 0 1 0 1
## 0110 6 0 1 1 0
## 0111 1 0 1 1 1
## 1000 5 1 0 0 0
## 1001 0 1 0 0 1
## 1010 0 1 0 1 0
## 1011 1 1 0 1 1
## 1100 15 1 1 0 0
## 1101 1 1 1 0 1
## 1110 6 1 1 1 0
## 1111 1 1 1 1 1
## attr(,"intersections")
## attr(,"intersections")$A
## [1] "18" "29" "32" "54" "58"
##
## attr(,"intersections")$B
## [1] "12" "14" "19" "22" "28" "30" "34" "36" "40" "43" "45" "46"
## [13] "51" "52" "53" "57" "60" "66" "67" "68" "69" "70" "73" "75"
## [25] "76" "77" "78" "80" "81" "87" "88" "89" "92" "94" "98" "100"
##
## attr(,"intersections")$C
## [1] "6" "10" "33" "84"
##
## attr(,"intersections")$D
## [1] "79"
##
## attr(,"intersections")$`A:B`
## [1] "2" "5" "9" "13" "17" "21" "25" "26" "27" "38" "39" "44" "49" "50" "56"
##
## attr(,"intersections")$`B:C`
## [1] "20" "71" "72" "82" "83" "97"
##
## attr(,"intersections")$`B:D`
## [1] "62" "63"
##
## attr(,"intersections")$`A:B:C`
## [1] "3" "11" "47" "74" "85" "95"
##
## attr(,"intersections")$`A:B:D`
## [1] "96"
##
## attr(,"intersections")$`A:C:D`
## [1] "7"
##
## attr(,"intersections")$`B:C:D`
## [1] "23"
##
## attr(,"intersections")$`A:B:C:D`
## [1] "37"
##
## attr(,"class")
## [1] "venn"
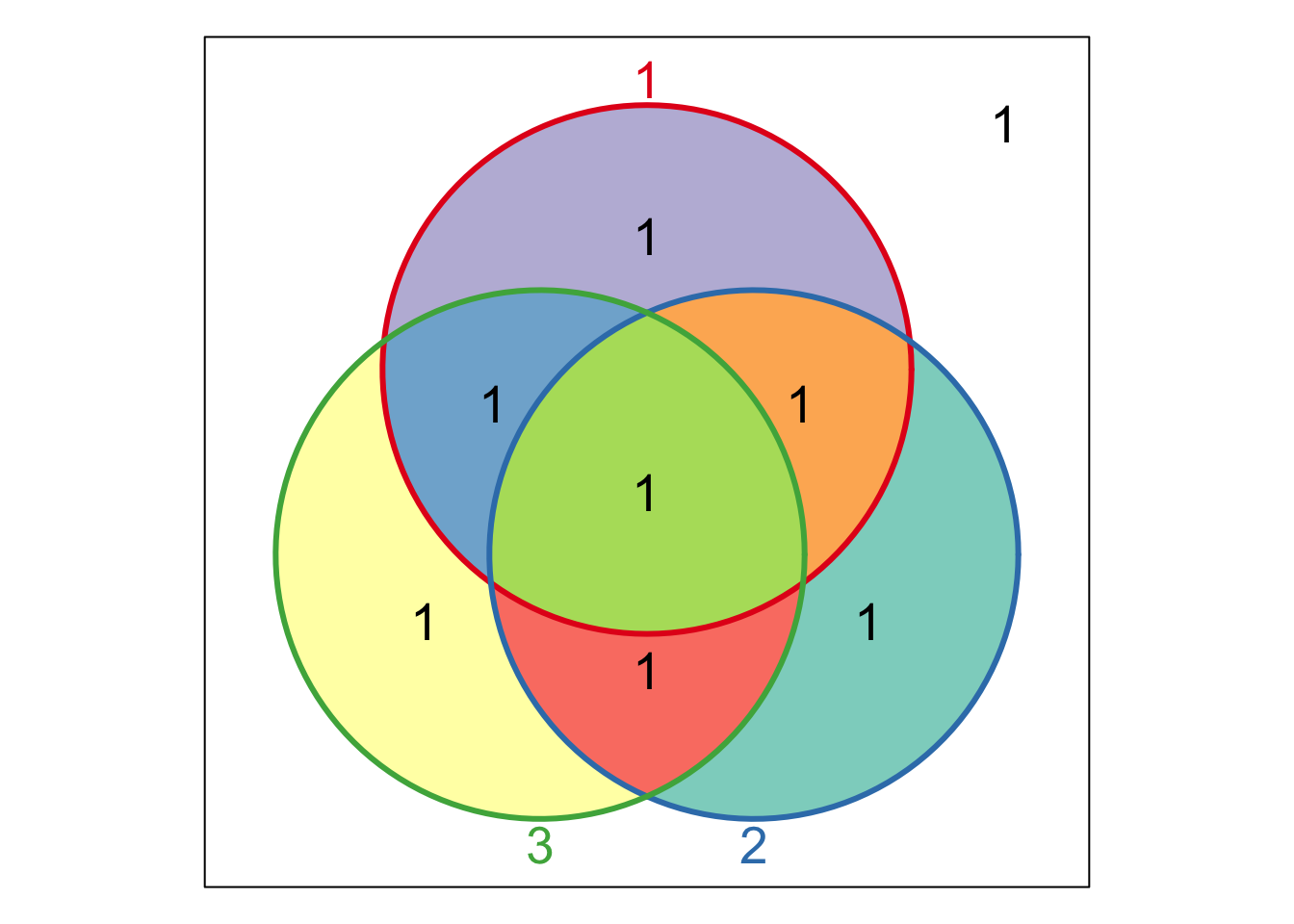
venn(input, showSetLogicLabel=TRUE)
# asbio包
library(asbio)
## 载入需要的程序包:tcltk
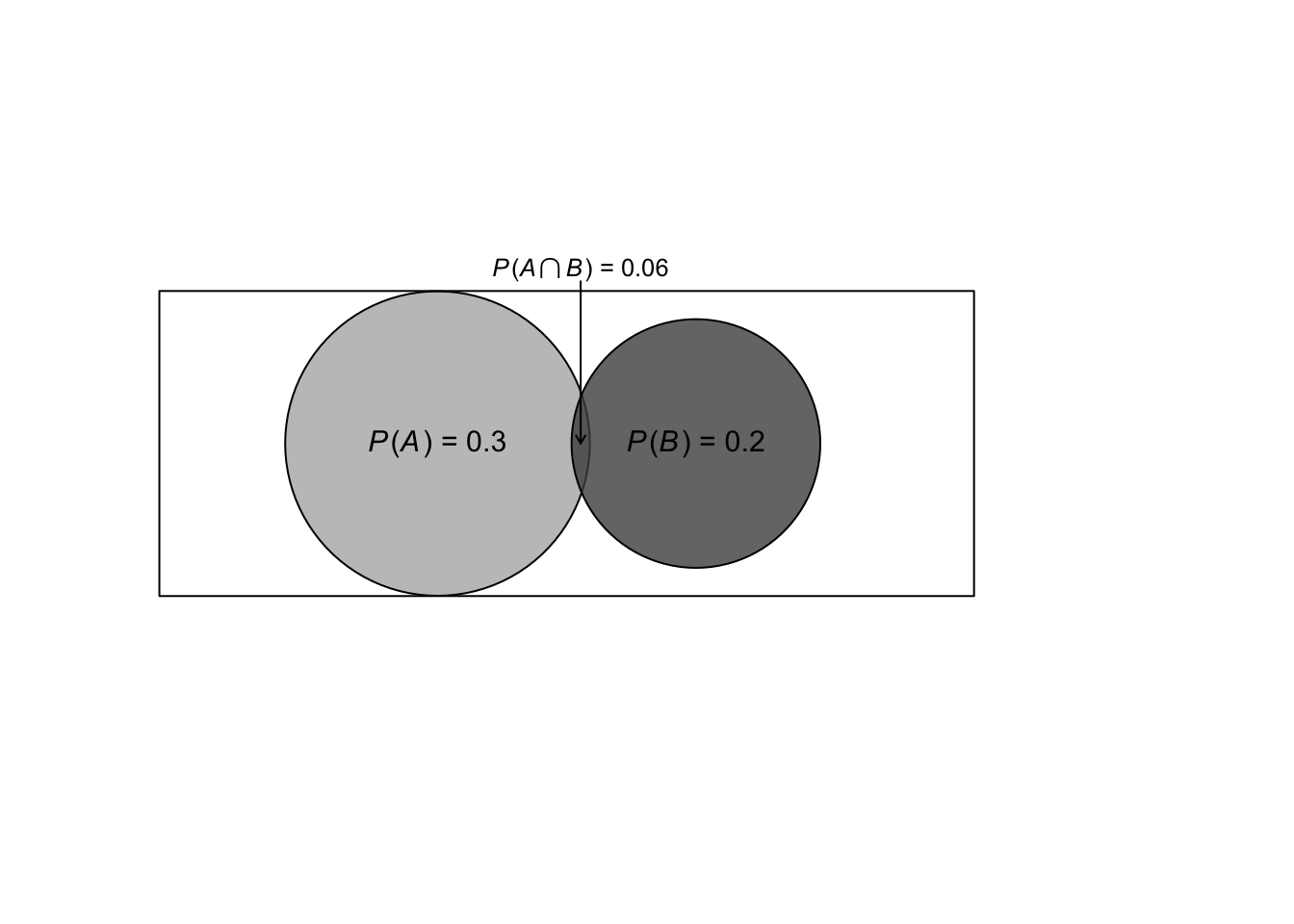
Venn(A=.3,B=.2,AandB=.06)
# limma包
# install.packages("BiocManager")
# BiocManager::install("limma")
library(limma)
Y <- matrix(rnorm(100*6),100,6)
Y[1:10,3:4] <- Y[1:10,3:4]+3
Y[1:20,5:6] <- Y[1:20,5:6]+3
design <- cbind(1,c(0,0,1,1,0,0),c(0,0,0,0,1,1))
fit <- eBayes(lmFit(Y,design))
results <- decideTests(fit)
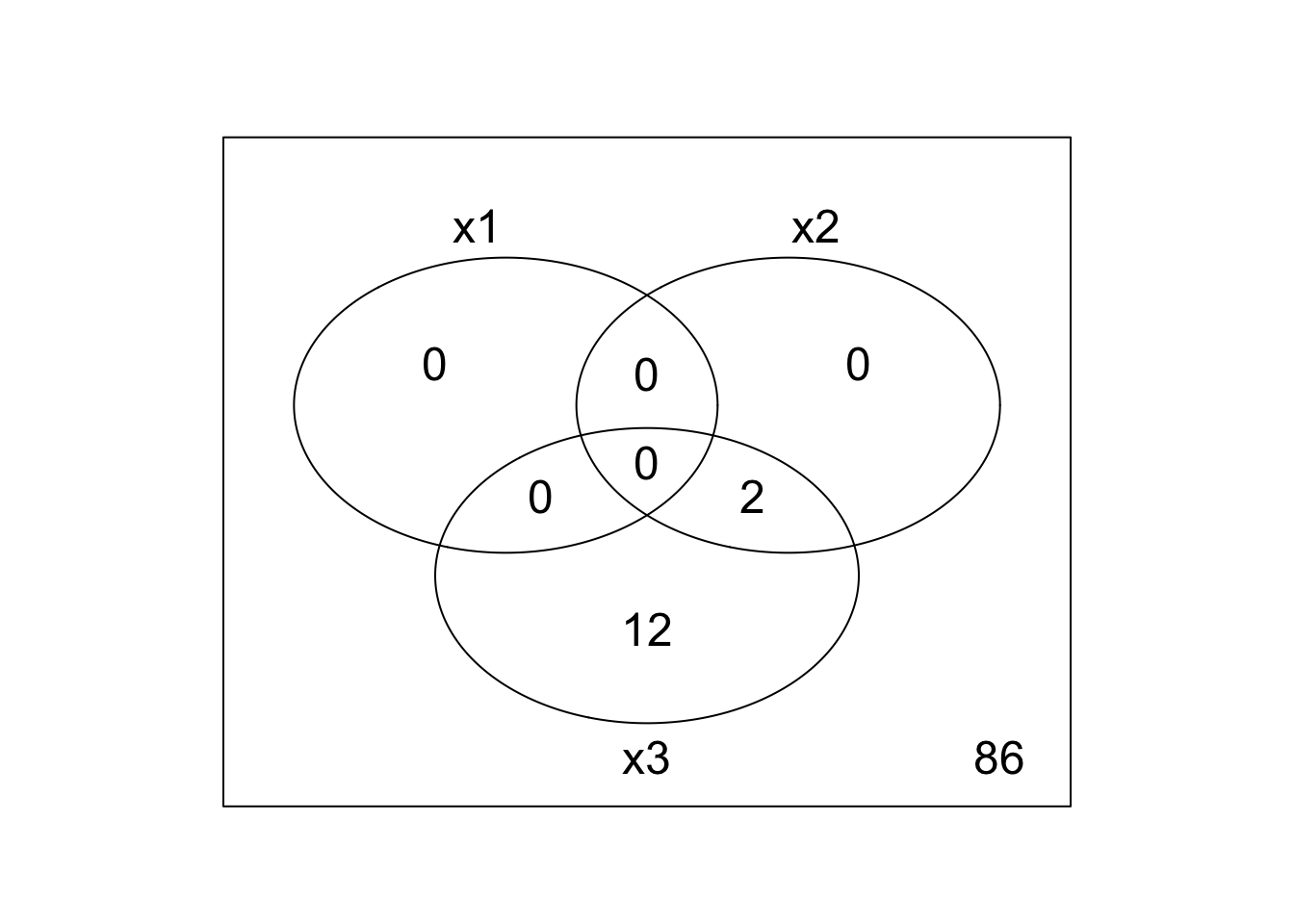
a <- vennCounts(results)
print(a)
## x1 x2 x3 Counts
## 1 0 0 0 88
## 2 0 0 1 8
## 3 0 1 0 0
## 4 0 1 1 4
## 5 1 0 0 0
## 6 1 0 1 0
## 7 1 1 0 0
## 8 1 1 1 0
## attr(,"class")
## [1] "VennCounts"
vennDiagram(a)
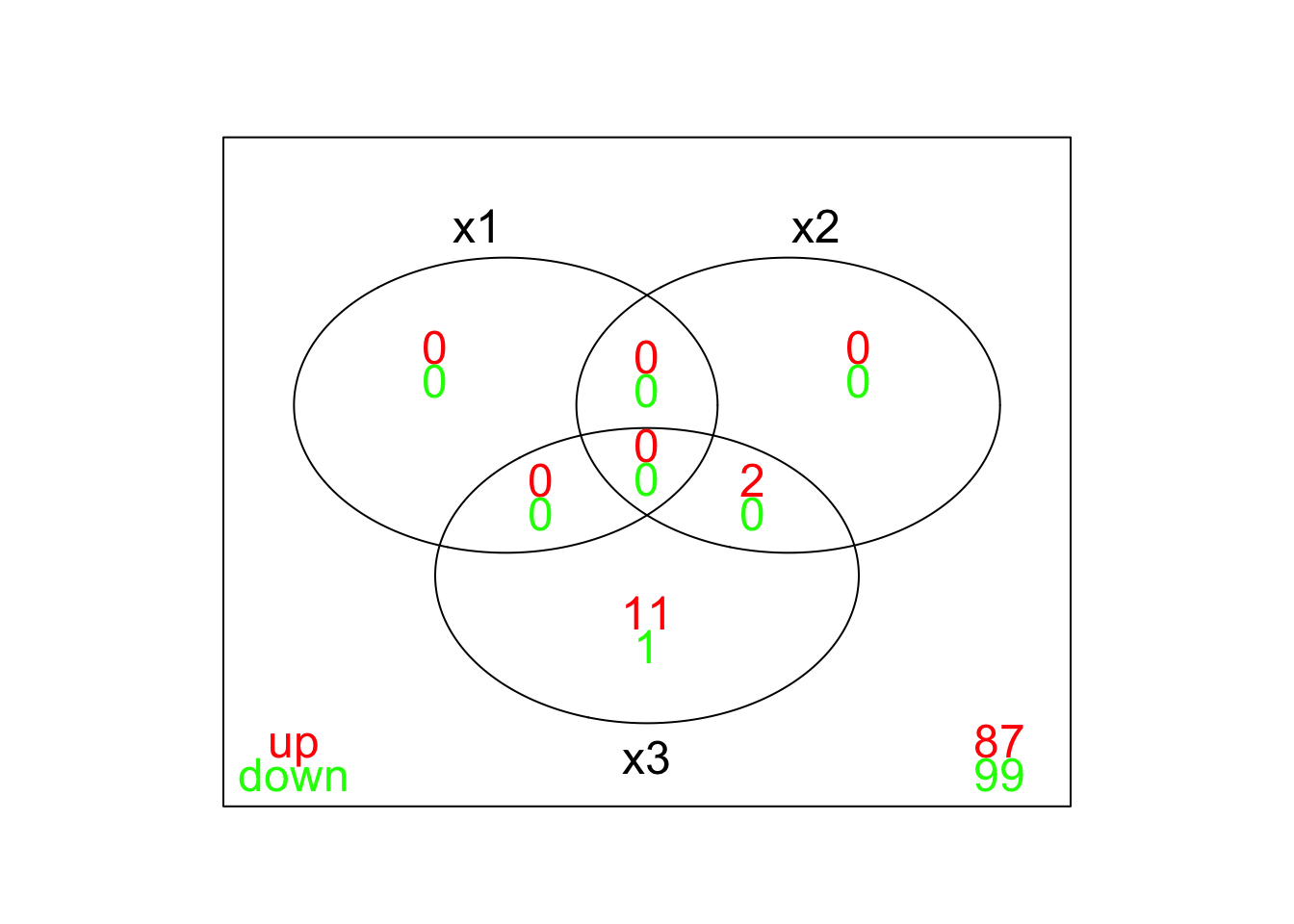
vennDiagram(results,include=c("up","down"),counts.col=c("red","green"))
# venneuler包
library(venneuler)
## 载入需要的程序包:rJava
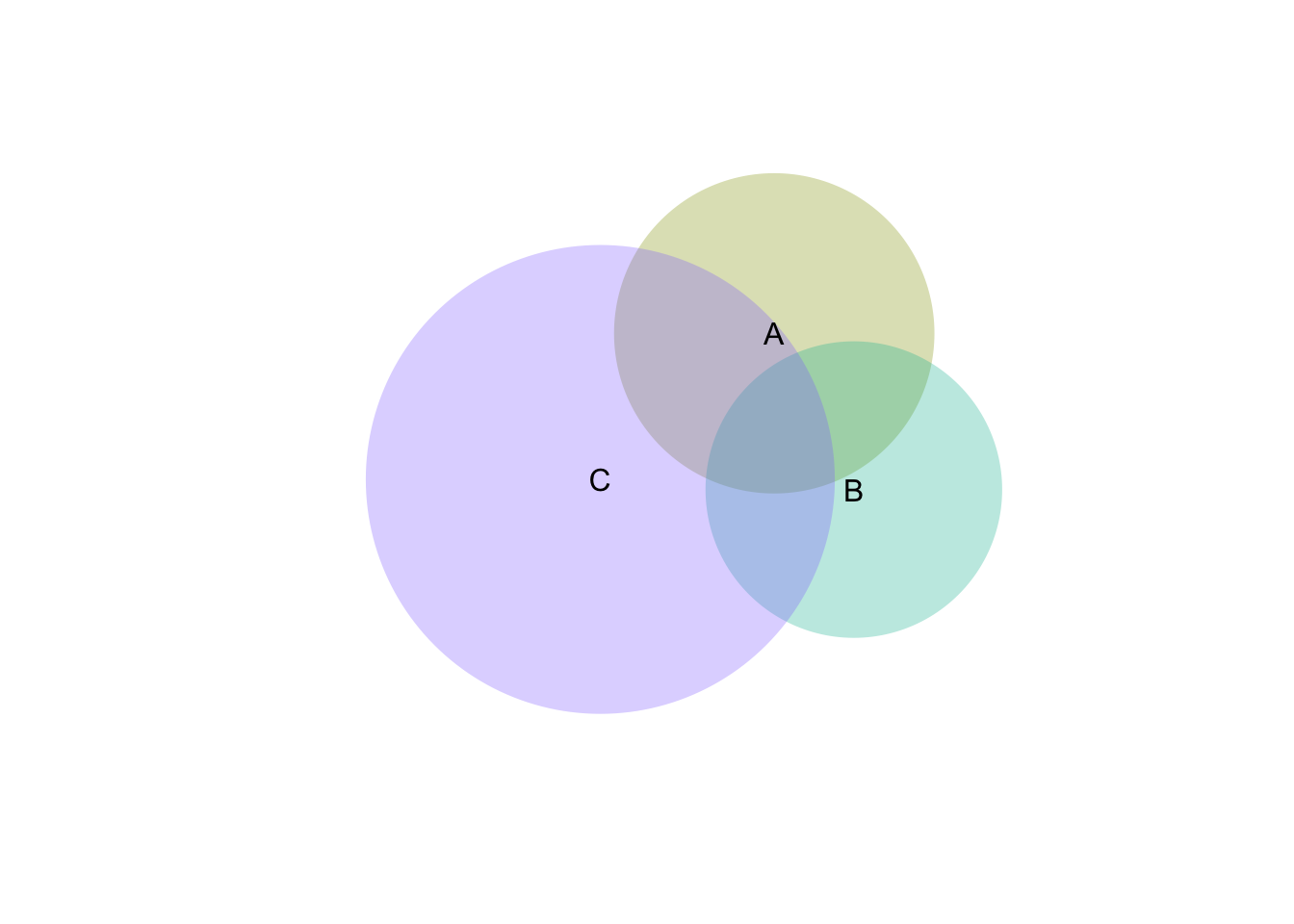
vd <- venneuler(c(A=0.3, B=0.3, C=1.1, "A&B"=0.1, "A&C"=0.2, "B&C"=0.1 ,"A&B&C"=0.1))
plot(vd)
m <- data.frame(elements=c("1","2","2","2","3"),sets=c("A","A","B","C","C"))
v <- venneuler(m)
plot(v)
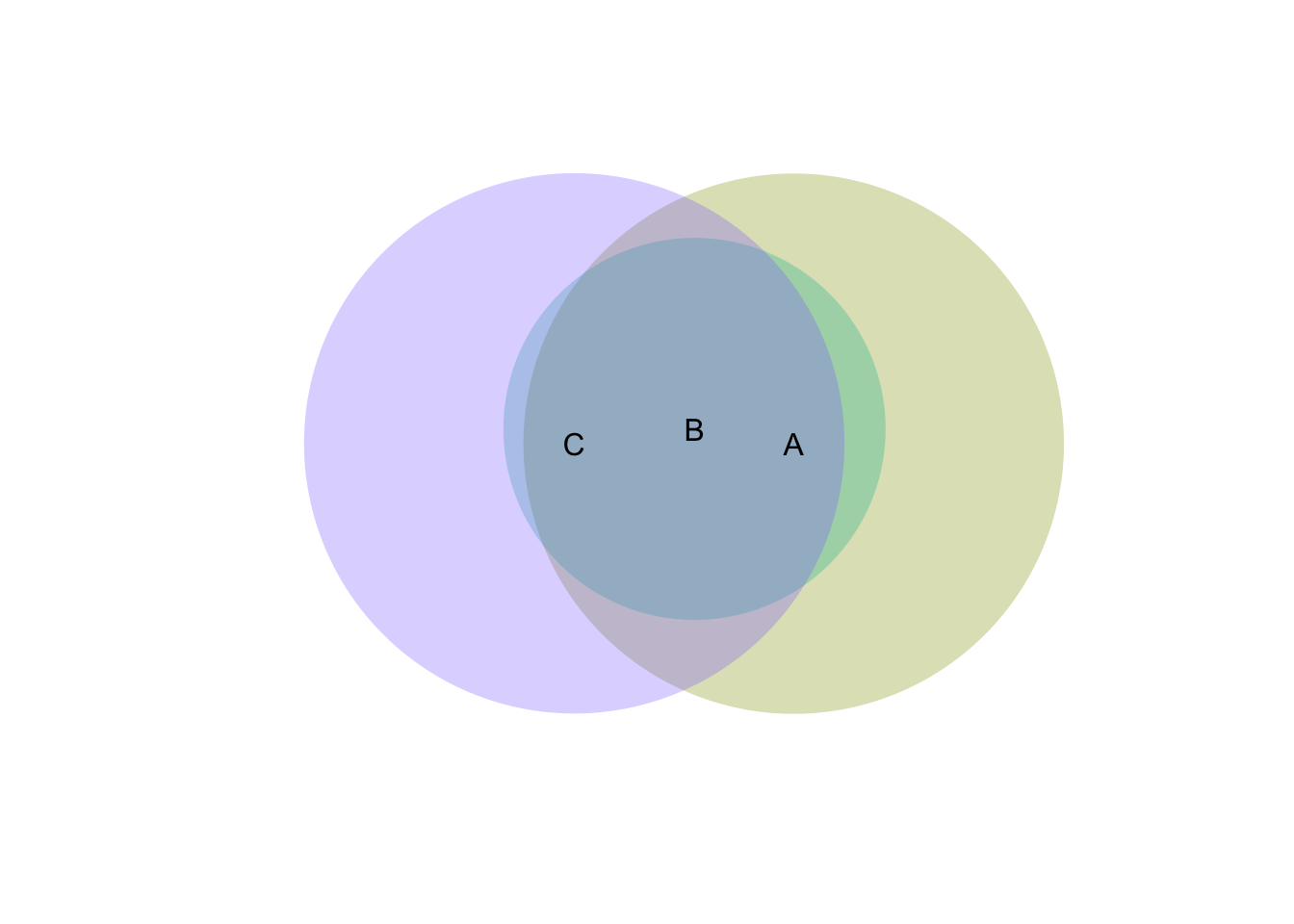
m <- as.matrix(data.frame(A=c(1.5, 0.2, 0.4, 0, 0),B=c(0, 0.2, 0, 1, 0), C=c(0, 0, 0.3, 0, 1)))
v <- venneuler(m > 0)
plot(v)
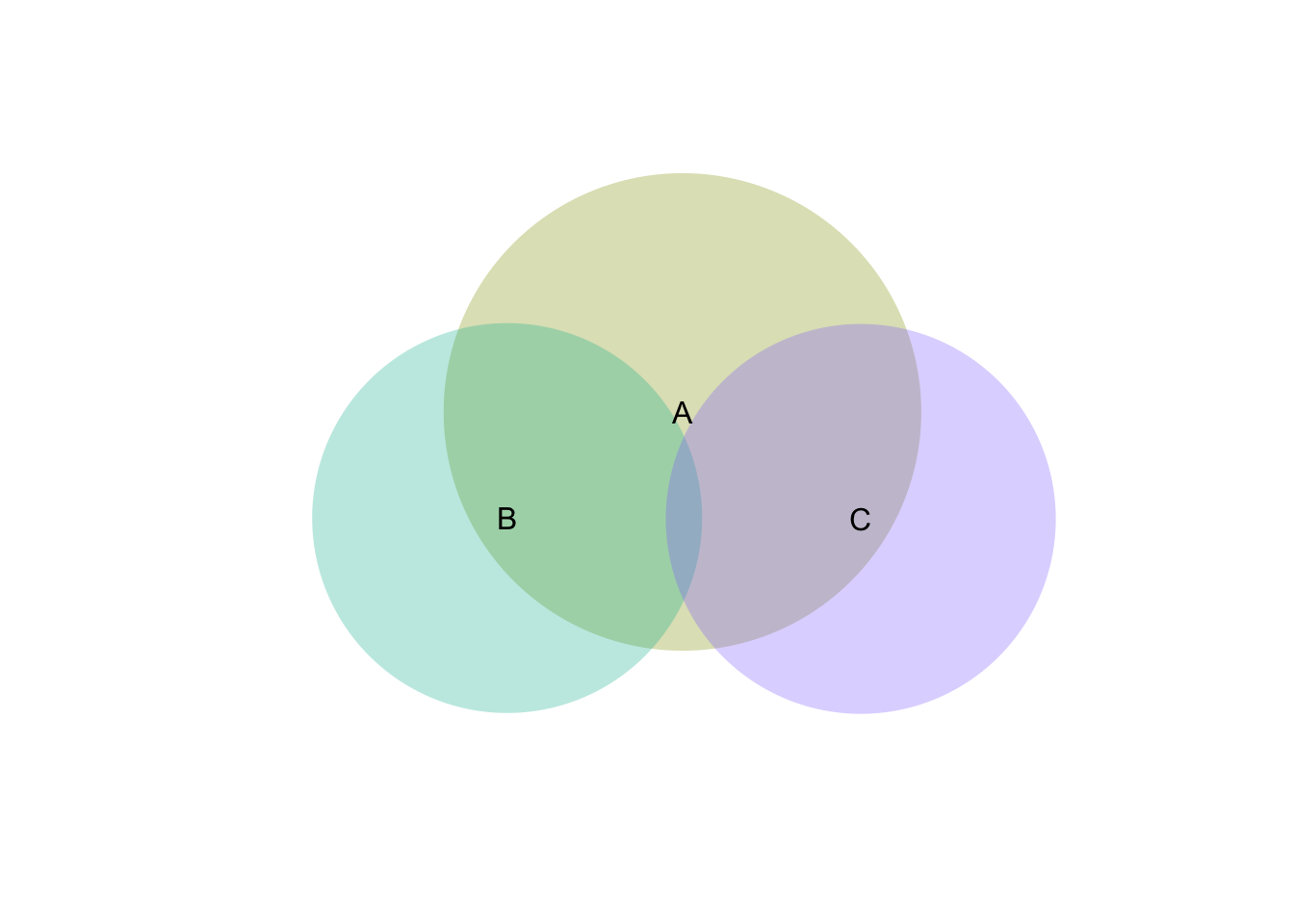
v <- venneuler(m)
plot(v)
# Vennerable包
# BiocManager::install("graph")
# BiocManager::install("RBGL")
# BiocManager::install("reshape")
# install.packages("Vennerable", repos="http://R-Forge.R-project.org")
library(Vennerable)
## 载入需要的程序包:graph
## 载入需要的程序包:BiocGenerics
## 载入需要的程序包:generics
##
## 载入程序包:'generics'
## The following objects are masked from 'package:base':
##
## as.difftime, as.factor, as.ordered, intersect, is.element, setdiff,
## setequal, union
##
## 载入程序包:'BiocGenerics'
## The following objects are masked from 'package:rJava':
##
## anyDuplicated, duplicated, unique
## The following object is masked from 'package:limma':
##
## plotMA
## The following objects are masked from 'package:stats':
##
## IQR, mad, sd, var, xtabs
## The following objects are masked from 'package:base':
##
## anyDuplicated, aperm, append, as.data.frame, basename, cbind,
## colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find,
## get, grep, grepl, is.unsorted, lapply, Map, mapply, match, mget,
## order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
## rbind, Reduce, rownames, sapply, saveRDS, table, tapply, unique,
## unsplit, which.max, which.min
## 载入需要的程序包:RBGL
## 载入需要的程序包:grid
## 载入需要的程序包:lattice
## 载入需要的程序包:RColorBrewer
## 载入需要的程序包:reshape
## 载入需要的程序包:gtools
## 载入需要的程序包:xtable
##
## 载入程序包:'Vennerable'
## The following object is masked from 'package:asbio':
##
## Venn
Venn(n=3)
## A Venn object on 3 sets named
## 1,2,3
## 000 100 010 110 001 101 011 111
## 1 1 1 1 1 1 1 1
plot(Venn(n=3))
data(StemCell)
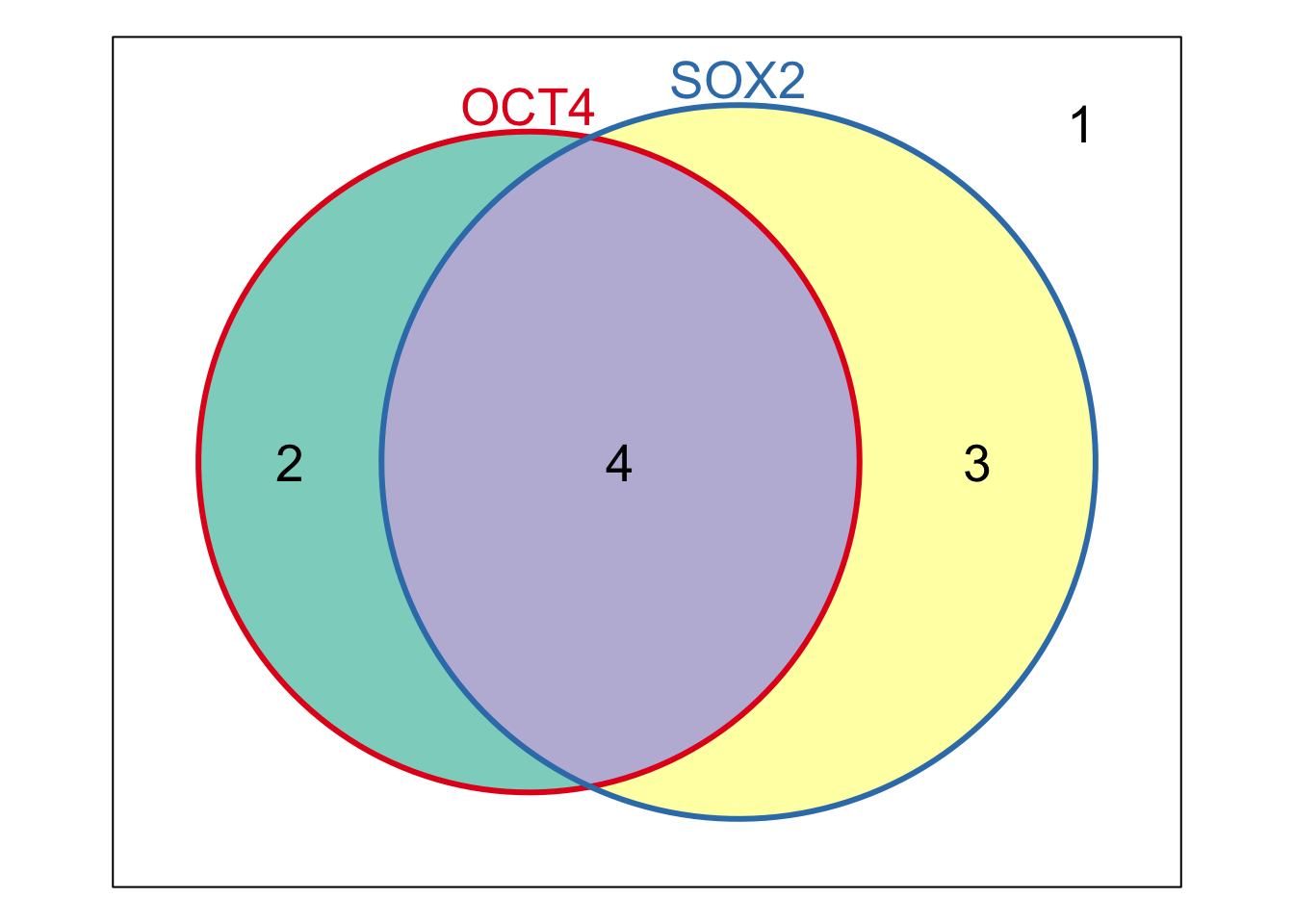
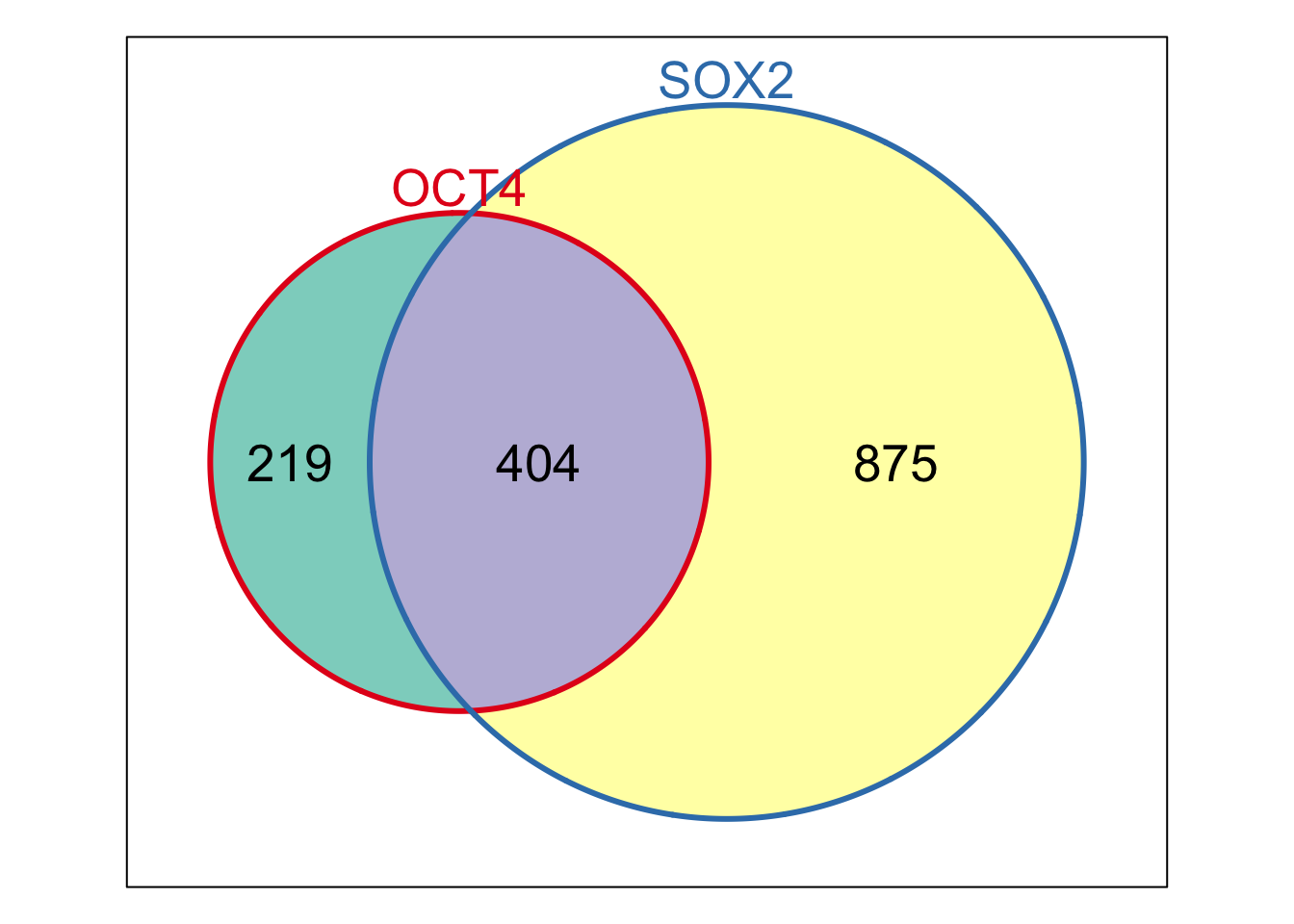
w <- Venn(Sets=StemCell[1:2])
Weights(w)
## 00 10 01 11
## 0 219 875 404
plot(w)
Weights(w) <- 1:4
plot(w)